I am an evolutionary and conservation genomicist investigating how genetic variation, from single nucleotide polymorphisms to large-scale structural changes, shapes adaptation across space and time. My research spans both invasive and native species, including plants and animals, to understand how genetic diversity is distributed throughout the genome, and how the diverse and complex forms of genetic variation underpin population resilience or vulnerability in the face of population disturbance or environmental change.
Structural Variants in Eco-evolutionary Genomics
- ❯ Structural variation in eco-evolutionary genomics (Stuart et al., 2025)
- ❯ Heterozygous structural variants uniquely impact lifetime fitness (Stuart et al., 2024)
- ❯ Population history shapes relative diversity levels between SVs and SNPs (Stuart et al., 2023a)

Evolutionary Genomics for Conservation & Restoration
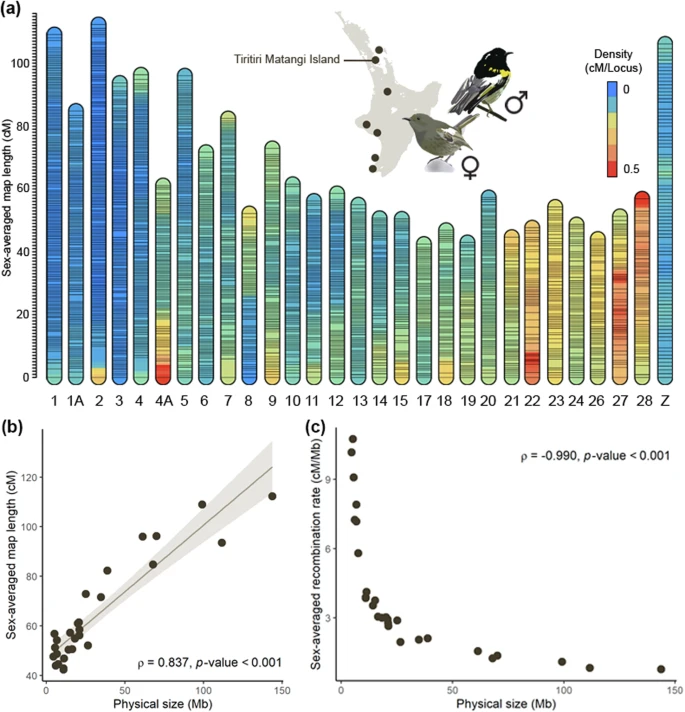
- ❯ Shaping diversity and evolutionary resilience in native seed production areas (forthcoming)
- ❯ Managing inbreeding within endangered Hihi populations (Tan et al., 2024) (Stuart et al., 2024a) (Tan et al., 2025)
Globally Invasive Common Starlings & Mynas
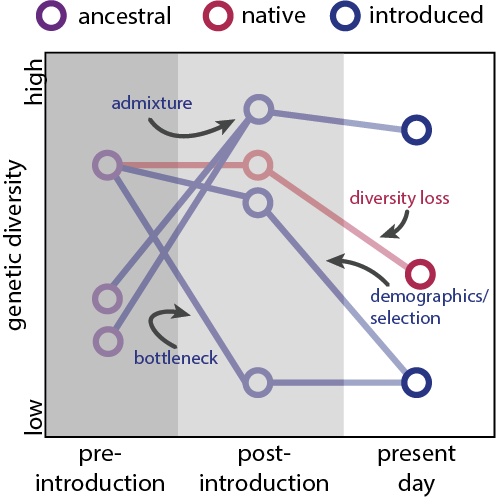
- ❯ Historical starling DNA from museum collections reveal global and invasive genomic changes during the Anthropocene (Stuart et al., 2022)
- ❯ Invasion driven change in a key domestication-associated gene shared between humans, dogs, sparrows, and invasive mynas (Atsawawaranunt et al., 2025)
- ❯ Historical newspaper records contextualize the starling invasion story told by DNA in New Zealand (Thompson et al., 2024)
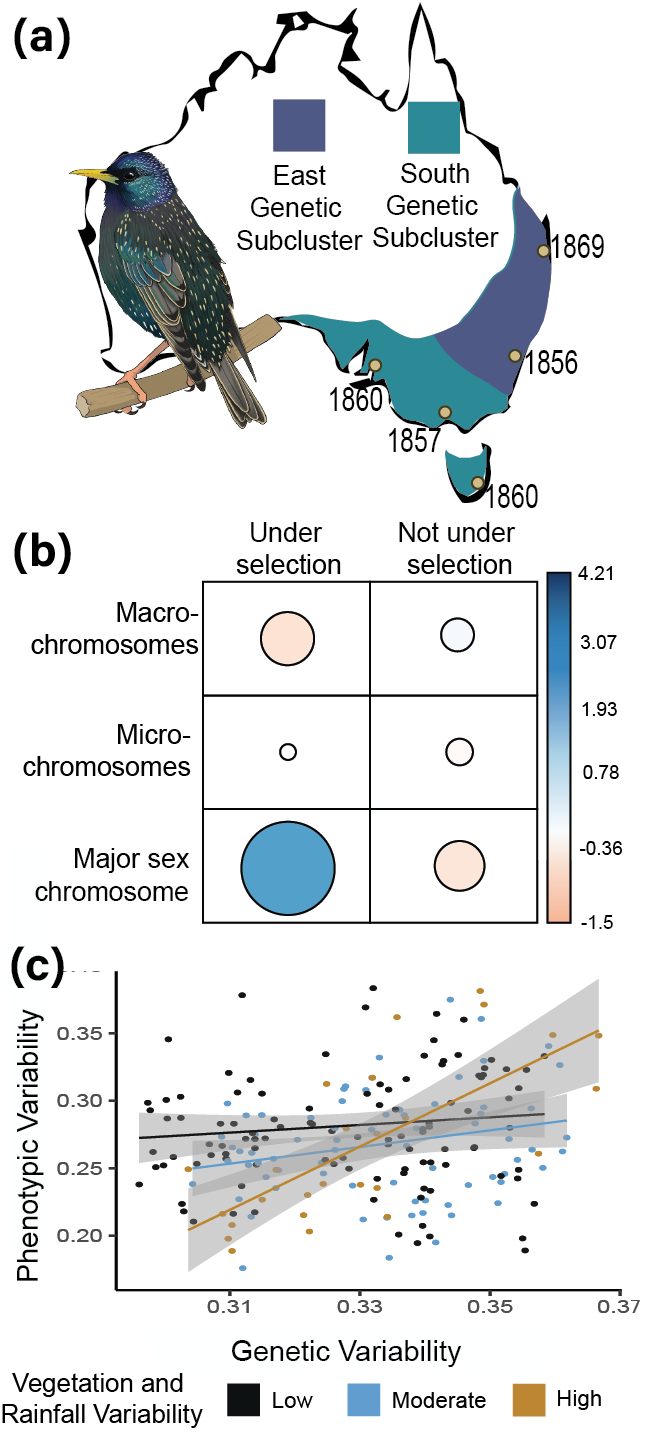
- ❯ Climate variability underlies morphological changes in Australia’s invasive starling populations (Stuart et al., 2023b)
Population Management for Invasive Species
- ❯ Genomic applications in invasion biology (McGaughran & Dhami et al. 2024) (Stuart et al. 2023c)
- ❯ Genomic-informed deer management in the Illawarra region (Li-Williams & Stuart et al., 2023)
Genomic diveristy & variation: Future research directions
❯ Detecting population genetic variation in eDNA.
❯ Population genomics of Australian native plants. This includes emerging work in seed production areas (SPAs), where we ask whether genetic diversity—neutral and functional—can predict restoration outcomes and population resilience.
